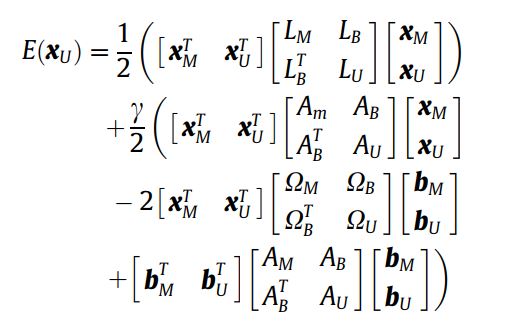Ошибка при использовании линейных решателей: ValueError: массив не должен содержать inf или NaN
Я получаю сообщение об ошибке при запуске кода.
ValueError: массив не должен содержать inf или NaN
Даже я пытался использовать x=np.nan_to_num(x) Решить проблему не удалось!
Примечание:
Код ниже - это настройка сегментации случайного обходчика из изображения scikit.
Приведенный ниже код является черновиком реализации бумажной сегментации путем извлечения с управляемыми случайными обходами.
Редактировать:
- Полная обратная связь:
Traceback (most recent call last):
File "D:\Saudi_CV\Vibot\Smester_2\2_Medical Image analysis\Project_2020\My_Classic_Project\My_Project\test.py", line 16, in <module>
img3 = random_walker(img, mask)
File "D:\Saudi_CV\Vibot\Smester_2\2_Medical Image analysis\Project_2020\My_Classic_Project\My_Project\Guided_RW.py", line 291, in random_walker
XL, XO, XA = solve_linear_system(lap_sparse,omega_sparse,A_sparse, B_L, B_O, B_A,tol)
File "D:\Saudi_CV\Vibot\Smester_2\2_Medical Image analysis\Project_2020\My_Classic_Project\My_Project\Guided_RW.py", line 177, in solve_linear_system
for i in range(B_O.shape[1])]
File "D:\Saudi_CV\Vibot\Smester_2\2_Medical Image analysis\Project_2020\My_Classic_Project\My_Project\Guided_RW.py", line 177, in <listcomp>
for i in range(B_O.shape[1])]
File "<decorator-gen-7>", line 2, in cg
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\_lib\_threadsafety.py", line 46, in caller
return func(*a, **kw)
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\sparse\linalg\isolve\iterative.py", line 317, in cg
work[slice1] = psolve(work[slice2])
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\sparse\linalg\interface.py", line 229, in matvec
y = self._matvec(x)
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\sparse\linalg\interface.py", line 527, in _matvec
return self.__matvec_impl(x)
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 314, in matvec
return self.solve(b, maxiter=1, cycle=cycle, tol=1e-12)
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 471, in solve
self.__solve(0, x, b, cycle)
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 518, in __solve
self.__solve(lvl + 1, coarse_x, coarse_b, 'V')
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 518, in __solve
self.__solve(lvl + 1, coarse_x, coarse_b, 'V')
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 518, in __solve
self.__solve(lvl + 1, coarse_x, coarse_b, 'V')
[Previous line repeated 4 more times]
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 515, in __solve
coarse_x[:] = self.coarse_solver(self.levels[-1].A, coarse_b)
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 711, in __call__
x = solve(self, A, b)
File "F:\Download_2019\Anaconda3\lib\site-packages\pyamg\multilevel.py", line 619, in solve
self.P = getattr(sp.linalg, solver)(A.todense(), **kwargs)
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\linalg\basic.py", line 1373, in pinv2
a = _asarray_validated(a, check_finite=check_finite)
File "F:\Download_2019\Anaconda3\lib\site-packages\scipy\_lib\_util.py", line 246, in _asarray_validated
a = toarray(a)
File "F:\Download_2019\Anaconda3\lib\site-packages\numpy\lib\function_base.py", line 499, in asarray_chkfinite
"array must not contain infs or NaNs")
ValueError: array must not contain infs or NaNs
Используемое изображение и маска здесь
Код, используемый для проверки алгоритма:
import numpy as np
import matplotlib.pyplot as plt
from Guided_RW import *
from skimage.io import imread
img = imread('img.png')
mask = imread('mask.png')
img3 = random_walker(img, mask)
plt.imshow(img3,cmap='gray')
plt.show()
import numpy as np
import time
import scipy
from scipy import sparse, ndimage as ndi
from scipy.sparse.linalg import cg, spsolve
from skimage import img_as_float
from distutils.version import LooseVersion as Version
import functools
if Version(scipy.__version__) >= Version('1.1'):
cg = functools.partial(cg, atol=0)
try:
from pyamg import ruge_stuben_solver
amg_loaded = True
except ImportError:
amg_loaded = False
def make_graph_edges(image):
if(len(image.shape)==2):
# print(image.shape)
n_x, n_y = image.shape
vertices = np.arange(n_x * n_y ).reshape((n_x, n_y))
edges_horizontal = np.vstack(( vertices[:, :-1].ravel(), vertices[:, 1:].ravel())) # X *(Y-1)
edges_vertical = np.vstack(( vertices[ :-1].ravel(), vertices[1: ].ravel())) #(X-1)* Y
edges = np.hstack((edges_horizontal, edges_vertical))
else:
print('image shape is: ',image.shape)
return edges
#===================================================================================
def compute_weights(image,mask,alpha, beta, eps=1.e-6):
intra_gradients = np.concatenate([np.diff(image, axis=ax).ravel()
for ax in [1, 0] ], axis=0) ** 2 # gradient ^2
# 5-Connected
inter_gradients = np.concatenate([np.diff(mask, axis=ax).ravel()
for ax in [1, 0] ], axis=0)**2
#----------------------------------------
# 1-Connected
# inter_gradients = (image - mask)**2
#----------------------------------------
# Normalize gradients
intra_gradients = (intra_gradients - np.amin(intra_gradients))/(np.amax(intra_gradients)- np.amin(intra_gradients))
inter_gradients = (inter_gradients - np.amin(inter_gradients))/(np.amax(inter_gradients)- np.amin(inter_gradients))
# All dimensions considered together in this standard deviation
#------------------------------------------------------
intra_scale_factor = -beta / (10 * image.std())
intra_weights = np.exp(intra_scale_factor * intra_gradients)
intra_weights += eps
#------------------------------------------------------
inter_scale_factor = -alpha / (10 * image.std())
inter_weights = np.exp(inter_scale_factor * inter_gradients)
inter_weights += eps
#------------------------------------------------------
return -intra_weights, inter_weights # [W_old , w_new]
#=============================================================================
def build_matrices(image, mask, alpha=90, beta=130):
edges_2D = make_graph_edges(image)
intra_weights, inter_weights = compute_weights(image=image,mask=mask,alpha=alpha ,beta=beta, eps=1.e-6 )
#================
# Matrix Laplace
#================
# Build the sparse linear system
pixel_nb = edges_2D.shape[1] # N = n_x * (n_y - 1) * + (n_x - 1) * n_y
i_indices = edges_2D.ravel() # Src - Dest
j_indices = edges_2D[::-1].ravel() # Same list in reverse order ( Dest - Src)
stacked_intra = np.hstack((intra_weights, intra_weights)) # weights (S-->D, D-->S) are same because graph is undirected
lap = sparse.coo_matrix((2*stacked_intra, (i_indices, j_indices)), shape=(pixel_nb, pixel_nb))
lap.setdiag(-2*np.ravel(lap.sum(axis=0)))
# print('Lap',lap.shape)
Laplace = lap.tocsr()
#================
# Matrix Omega
#================
# Build the sparse linear system
stacked_inter = np.hstack((inter_weights, inter_weights)) # weights (S-->D, D-->S) are same because graph is undirected
Omeg = sparse.coo_matrix((2*stacked_inter, (i_indices, j_indices)), shape=(pixel_nb, pixel_nb))
Omeg.setdiag(2*np.ravel((image-mask)**2))
# print('Omeg',Omeg.shape)
Omega = Omeg.tocsr()
#================
# Matrix A
#================
# Build the sparse linear system
weights = Omega.copy()
firstColumn = weights.sum(axis=1)/2
otherColumns = sparse.csr_matrix((weights.shape[0],weights.shape[1]-1))
Mat_A = sparse.hstack((firstColumn, otherColumns))
# print('A',Mat_A.shape)
return Laplace, Omega, Mat_A
#=============================================================================
def build_linear_system(image, labels, nlabels, beta):
"""
Build the matrix A and rhs B of the linear system to solve.
A and B are two blocks of the laplacian of the image graph.
"""
labels2 = labels.copy()
labels = labels.ravel()
indices = np.arange(labels.size) # indices of all vertices
seeds_mask = labels > 0 # Keep only lables withL_out Background
unlabeled_indices = indices[~seeds_mask] # U
seeds_indices = indices[seeds_mask] # M
lap_sparse,omega_sparse, A_sparse = build_matrices(image=image, mask=labels2,
beta=beta)
A_sparse = A_sparse.tocsr()
#-------------------------------------------------------------------------
seeds = labels[seeds_mask]
seeds_mask = sparse.csc_matrix(np.hstack([np.atleast_2d(seeds == lab).T
for lab in range(1, nlabels + 1)]))
#-------------------------------------------------------------------------
# Laplacian
#=============
rows_L = lap_sparse[unlabeled_indices, :]
lap_sparse = rows_L[:, unlabeled_indices] # size = U x U
B_L = -rows_L[:, seeds_indices] # size = N^2(U+M) x U
rhs_L = B_L.dot(seeds_mask)
#-------------------------------------------------------------------------
# Omega
#=============
rows_O = omega_sparse[unlabeled_indices, :]
omega_sparse = rows_O[:, unlabeled_indices] # size = U x U
B_O = -rows_O[:, seeds_indices] # size = N^2(U+M) x U
rhs_O = B_O.dot(seeds_mask)
#-------------------------------------------------------------------------
rows_A = A_sparse[unlabeled_indices, :]
A_sparse = rows_A[:, unlabeled_indices] # size = U x U
B_A = -rows_A[:, seeds_indices] # size = N^2(U+M) x U
rhs_A = B_A.dot(seeds_mask)
#-------------------------------------------------------------------------
return lap_sparse, omega_sparse, A_sparse, rhs_L, rhs_O, rhs_A
#=============================================================================
def solve_linear_system(lap_sparse,omega_sparse,A_sparse, B_L, B_O, B_A,tol):
if not amg_loaded:
print('"cg_mg" not available, it requires pyamg to be installed. ',
'The "cg_j" mode will be used instead.')
#-------------------------------------------------------------------------------
# Laplace
#==========
maxiter = 30
lap_sparse = lap_sparse.tocsr()
mlL = ruge_stuben_solver(lap_sparse)
ML = mlL.aspreconditioner(cycle='V')
cg_L_out_L = [
cg(lap_sparse, B_L[:, i].toarray(), tol=tol, M=ML, maxiter=maxiter)
for i in range(B_L.shape[1])]
if np.any([info > 0 for _, info in cg_L_out_L]):
print("Laplace Conjugate gradient convergence to tolerance not achieved. ",
"\nConsider decreasing beta to improve system conditionning.")
XL = np.asarray([x1 for x1, _ in cg_L_out_L])
#-------------------------------------------------------------------------------
# Omega
#==========
omega_sparse = omega_sparse.tocsr()
mlO = ruge_stuben_solver(omega_sparse)
MO = mlO.aspreconditioner(cycle='V')
maxiter = 30
B_O = np.nan_to_num(B_O)
omega_sparse = np.nan_to_num(omega_sparse)
cg_L_out_O = [
cg(omega_sparse, B_O[:, i].toarray(), tol=tol, M=MO, maxiter=maxiter)
for i in range(B_O.shape[1])]
if np.any([info > 0 for _, info in cg_L_out_O]):
print("Omega Conjugate gradient convergence to tolerance not achieved. ",
"\nConsider decreasing beta to improve system conditionning.")
XO = np.asarray([x1 for x1, _ in cg_L_out_O])
#--------------------------------------------------------------------------------
# Mat_A
#==========
A_sparse = A_sparse.tocsr()
mlA = ruge_stuben_solver(A_sparse)
MA = mlA.aspreconditioner(cycle='V')
maxiter = 30
cg_L_out_A = [
cg(A_sparse, B_A[:, i].toarray(), tol=tol, M=MA, maxiter=maxiter)
for i in range(B_A.shape[1])]
if np.any([info > 0 for _, info in cg_L_out_A]):
print("Mat_A Conjugate gradient convergence to tolerance not achieved. ",
"\nConsider decreasing beta to improve system conditionning.")
XA = np.asarray([x1 for x1, _ in cg_L_out_A])
#--------------------------------------------------------------------------------
return XL, XO, XA
#=======================================================================
def preprocess(labels):
label_values, inv_idx = np.unique(labels, return_inverse=True)
if not (label_values == 0).any():
print('Random walker only segments unlabeled areas, where ',
'\nlabels == 0. No zero valued areas in labels were ',
'\nfound. Returning provided labels.')
return labels, None, None, None, None
# If some labeled pixels are isolated inside pruned zones, prune them
# as well and keep the labels for the final L_output
null_mask = labels == 0
pos_mask = labels > 0
mask = labels >= 0
fill = ndi.binary_propagation(null_mask, mask=mask)
isolated = np.logical_and(pos_mask, np.logical_not(fill))
pos_mask[isolated] = False
# If the array has pruned zones, be sure that no isolated pixels
# exist between pruned zones (they could not be determined)
if label_values[0] < 0 or np.any(isolated):
isolated = np.logical_and(
np.logical_not(ndi.binary_propagation(pos_mask, mask=mask)),
null_mask)
labels[isolated] = -1
if np.all(isolated[null_mask]):
print('All unlabeled pixels are isolated, they could not be ',
'\ndetermined by the random walker algorithm.')
return labels, None, None, None, None
mask[isolated] = False
mask = np.atleast_2d(mask)
else:
mask = None
# Reorder label values to have consecutive integers (no gaps)
zero_idx = np.searchsorted(label_values, 0)
labels = np.atleast_2d(inv_idx.reshape(labels.shape) - zero_idx)
nlabels = label_values[zero_idx + 1:].shape[0]
inds_isolated_seeds = np.nonzero(isolated)
isolated_values = labels[inds_isolated_seeds]
return labels, nlabels, mask, inds_isolated_seeds, isolated_values
#=========================================================================
def random_walker(image, labels, beta=130, gamma=0.4, tol=1.e-3, copy=True, return_full_prob=False):
if (image.ndim !=2):
raise ValueError('For non-multichannel input, image must be of '
'dimension 2.')
if image.shape != labels.shape:
raise ValueError('Incompatible image and labels shapes.')
# image = np.atleast_2d(img_as_float(image))[..., np.newaxis]
image = img_as_float(image)#
labels_shape = labels.shape
labels_dtype = labels.dtype
if copy:
labels = np.copy(labels)
(labels, nlabels, mask,
inds_isolated_seeds, isolated_values) = preprocess(labels)
if isolated_values is None:
# No non isolated zero valued areas in labels were
# found. Returning provided labels.
if return_full_prob:
# Return the concatenation of the masks of each unique label
return np.concatenate([np.atleast_2d(labels == lab)
for lab in np.unique(labels) if lab > 0],
axis=-1)
return labels
# Build the linear system (lap_sparse, B)
lap_sparse,omega_sparse,A_sparse,B_L,B_O,B_A = build_linear_system(image,
labels, nlabels, beta)
# Solve the linear system lap_sparse X = B
# where X[i, j] is the probability that a marker of label i arrives
# first at pixel j by anisotropic diffusion.
XL, XO, XA = solve_linear_system(lap_sparse,omega_sparse,A_sparse, B_L, B_O, B_A,tol)
# Build the L_output according to return_full_prob value
# Put back labels of isolated seeds
labels[inds_isolated_seeds] = isolated_values
labels = labels.reshape(labels_shape)
#-----------------------------------------------------------------------------
if return_full_prob:
mask = labels == 0
#===========#
# Laplacian #
#===========#
L_out = np.zeros((nlabels,) + labels_shape)
for lab, (label_prob_L, prob_L) in enumerate(zip(L_out, XL), start=1):
label_prob_L[mask] = prob_L
label_prob_L[labels == lab] = 1
#===========#
# Omega #
#===========#
O_out = np.zeros((nlabels,) + labels_shape)
for lab, (label_prob_O, prob_O) in enumerate(zip(O_out, XO), start=1):
label_prob_O[mask] = prob_O
label_prob_O[labels == lab] = 1
#===========#
# Mat_A #
#===========#
A_out = np.zeros((nlabels,) + labels_shape)
for lab, (label_prob_A, prob_A) in enumerate(zip(O_out, XO), start=1):
label_prob_A[mask] = prob_O
label_prob_A[labels == lab] = 1
#-----------------------------------------------------------------------------
else:
#===========#
# Laplacian #
#===========#
XL = np.argmax(XL, axis=0) + 1
L_out = labels.astype(labels_dtype)
L_out[labels == 0] = XL
#===========#
# Omega #
#===========#
XO = np.argmax(XO, axis=0) + 1
O_out = labels.astype(labels_dtype)
O_out[labels == 0] = XO
#===========#
# Mat_A #
#===========#
XA = np.argmax(XA, axis=0) + 1
A_out = labels.astype(labels_dtype)
A_out[labels == 0] = XA
#-----------------------------------------------------------------------------
return 0.5*L_out + 0.5*gamma*A_out - 2*O_out